Genome Sequencing at the Culture Collection of Algae and Protozoa (CCAP)
Thursday 6 February
The CCAP holds a uniquely wide diversity of microalgae, protists and seaweeds. In 2019, we progressed with sequencing the genomes of a range of our microbial strains. The Collection was invited to join two external projects that aim to sequence the whole genomes of eukaryotic algae and protists: the Darwin Tree of Life Project and the 10KP Whole Genome Sequencing Project. Within CCAP we undertook the whole genome sequencing of 34 cyanobacterial cultures.
Our approach for CCAP’s cyanobacteria was to employ high-throughput metagenomic sequencing and exploit recent advances in the quality, sequencing depth and reduced costs of next-generation DNA sequencers. All cyanobacteria were successfully assembled to >90 % completion (Fig. 1), along with a further 431 bacterial genomes that represent the heterotrophic bacterial communities (aka microbiome) found living with the Cyanobacteria.

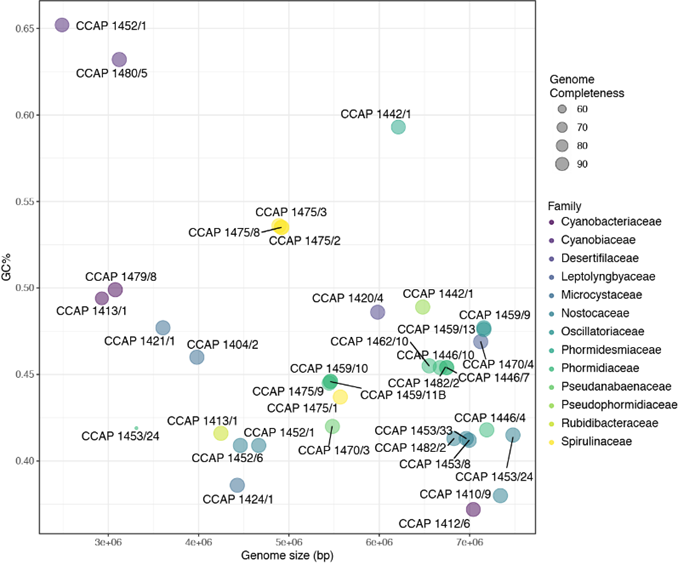
Figure 1: Size and GC% of the CCAP genome-assembled Cyanobacteria. Each strain is coloured according to their taxonomic group and genome completeness. GTDB-TK and CheckM were used to infer taxonomy and genome completion, respectively.
CCAP is preparing a publication reporting the high quality cyanobacterial assembly and whole genome-based taxonomy using the new metagenome assembly methods. This work is an important proof-of-principal that, for relatively low costs, we can now reliably assemble heavily contaminated cyanobacterial cultures and avoid the fate of some current cyanobacterial genomes which are badly contaminated with associated bacteria.
Further analysis is investigating the taxonomic composition and functional contribution of the assembled microbiomes, addressing how cyanobacterial biogeography, taxonomy and genotype affect the composition of their microbiomes. Preliminary evidence shows that the microbiomes taxonomic composition is strongly affected by whether cultures live in saline or freshwater/terrestrial environments (Fig. 2).


Figure 2. Salinity is a strong driver of cyanobacterial microbiome composition. Non-metric multidimensional scaling (nMDS) of the Bray-Curtis distance between communities calculated from the abundance of bacterial taxa between cyanobacterial cultures. Each microbiome is coloured according to the biome it was originally isolated from.